Program 11: EMS Stations
CSci 39542: Introduction to Data Science
Department of Computer Science
Hunter College, City University of New York
Spring 2022
Classwork Quizzes Homework Project
Program Description
Program 11: EMS Stations. Due 10am, Wednesday, 26 April.
For this program, we are building on Classwork 10
which focused ambulance calls in New York City.
Decreasing ambulance response times improves outcomes and
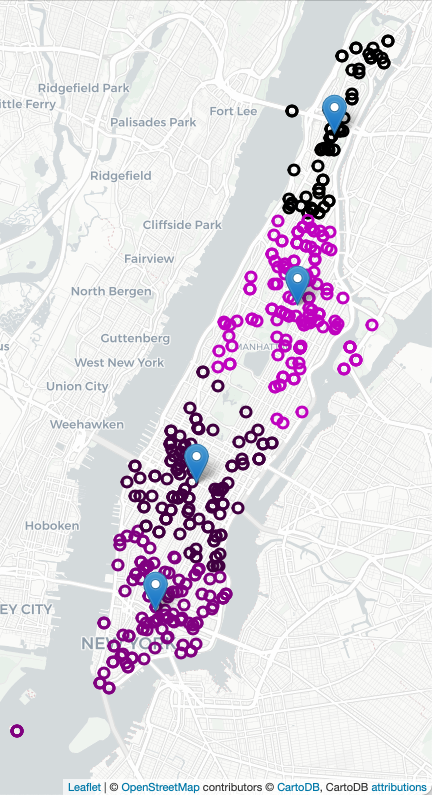
strategic placement of ambulance stations and overall allocation has been shown an effective approach. For example, here are all the calls for ambulances on 4 July 2021 in Manhattan (using Folium/Leaflet to create an interactive map):
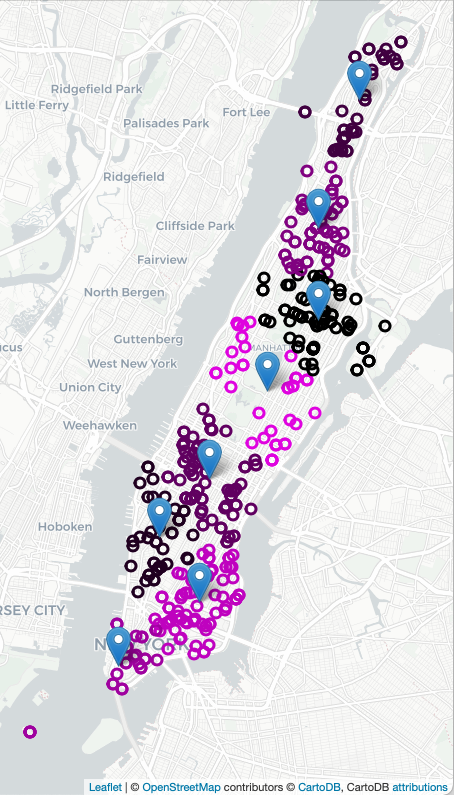
To decide on where to "pre-place" ambulances, we will use K-means clustering, where "K" is the number of ambulances available for that shift. For example, if there 2 ambulances available to be placed in Manhattan, we will look at previous ambulance calls for that shift and form 2 clusters and station each ambulance at the mean of the cluster. If two more ambulances become available, we can recompute the K-means algorithm for K=4, and place those 4 ambulances, each at the mean of the cluster found, and similarly for K=8:
In addition to K-means analysis, we will also use standard approaches from sklearn as well as filter data by time and day.
Learning Objective: to enhance data cleaning skills and build understanding of clustering algorithms.
Available Libraries: pandas, datetime, numpy, sklearn, and core Python 3.6+.
Data Sources: 911 System Calls (NYC OpenData)
Sample Datasets:

The assignment is broken into the following functions to allow for unit testing:
-
make_df(file_name): This function takes one input:-
file_name: the name of a CSV file containing 911 System Calls from OpenData NYC.
AMBULANCEas part of theTYP_DESCare kept. The resulting DataFrame is returned.
Hint: see DS 100: Chapter 13 for using string methods within pandas.
See Classwork 9.
-
-
add_date_time_features(df): This function takes one input:-
df: a DataFrame containing 911 System Calls from OpenData NYC created bymake_df.
WEEK_DAYis added with the day of the week (0 for Monday, 1 for Tuesday, ..., 6 for Sunday) of the date inINCIDENT_DATEis added. Another column,INCIDENT_MIN, that takes the time fromINCIDENT_TIMEand stores it as the number of minutes since midnight. The resulting DataFrame is returned.
Hint: see Lecture 3 for using datetime methods with pandas, including computing the day of the week (ofdatetimeobjects) and the total seconds (oftimedeltaobjects).
-
-
filter_by_time(df, days=None, start_min=0, end_min=1439): This function takes four inputs:-
df: a DataFrame containing 911 System Calls from OpenData NYC. -
days: a list of integers ranging from 0 to 6, representing the days of the week. The default value isNoneand is equivalent to the list containing all days:[0,1,2,3,4,5,6]. -
start_min: a non-negative integer value representing the starting time. Withend_min, it representing the range, inclusive, for the time, in minutes, that should be selected. The default value give the range of[0,1439]which ranges from midnight (0minutes) to (1439representing 23:59 since 23 hours + 59 minutes = 23*60+59 minutes = 1439 minutes). -
end_min: a non-negative integer value representing the ending time. Withstart_min, it representing the range, inclusive, for the time, in minutes, that should be selected. The default value give the range of[0,1439]which ranges from midnight (0minutes) to (1439representing 23:59 since 23 hours + 59 minutes = 23*60+59 minutes = 1439 minutes).
days(or all weekdays ifNoneis given) and incident times in[start_min, end_min]inclusive (e.g. contains the endpoints).
-
-
compute_kmeans(df, num_clusters = 8, n_init = 'auto', random_state = 2022): This function takes four inputs:-
df: a DataFrame containing 911 System Calls from OpenData NYC. -
n_init: Number of times the k-means algorithm is run with different centroid seeds. The final results is the best output ofn_initconsecutive runs in terms of inertia. The default value isauto. -
num_clusters: an integer representing the number of clusters. The default value is8. -
random_state: the random seed used for KMeans. The default value is2022.
num_clusterson the latitude and longitude data of the provided DataFrame. Returns the cluster centers and predicted labels computed via the model.
A similar, but not identical function was part of Classwork 9.
-
-
compute_gmm(df, num_clusters = 8, random_state = 2022): This function takes three inputs:-
df: a DataFrame containing 911 System Calls from OpenData NYC. -
num_clusters: an integer representing the number of clusters. The default value is8. -
random_state: the random seed used for GaussianMixture. The default value is2022.
num_clusterson the latitude and longitude data of the provided DataFrame. Returns the array of the predicted labels computed via the model.
-
-
compute_agglom(df, num_clusters = 8, linkage='ward'): This function takes three inputs:-
df: a DataFrame containing 911 System Calls from OpenData NYC. -
num_clusters: an integer representing the number of clusters. The default value is8. -
linkage: the linkage criterion used determining distances between sets for AgglomerativeClustering. The default value is'ward'.
num_clusterson the latitude and longitude data of the provided DataFrame and default linkage (i.e.ward). Returns the array of the predicted labels computed via the model.
-
-
compute_explained_variance(df, K =[1,2,3,4,5], random_state = 55): This function takes three inputs:-
df: a DataFrame containing 911 System Calls from OpenData NYC. -
K: a list of integers representing values for the number of clusters. The default value is[1,2,3,4,5]. -
random_state: the random seed used for KMeans. The default value is55.
K. This can be computed manually or via theinertia_attribute of the KMeans model. -
For example, if we use the small dataset from 4 July 2021:
df = make_df('NYPD_Calls_Manhattan_4Jul2021.csv')
print(df[['INCIDENT_TIME','TYP_DESC','Latitude','Longitude']])
INCIDENT_TIME TYP_DESC Latitude Longitude
7 00:01:51 AMBULANCE CASE: CARDIAC/OUTSIDE 40.724578 -73.992519
27 00:06:12 AMBULANCE CASE: CARDIAC/INSIDE 40.807719 -73.964240
51 00:12:12 AMBULANCE CASE: SERIOUS/TRANSIT 40.732019 -74.000734
53 00:12:38 AMBULANCE CASE: EDP/INSIDE 40.789348 -73.947352
54 00:12:38 AMBULANCE CASE: EDP/INSIDE 40.789348 -73.947352
... ... ... ... ...
5175 23:50:02 AMBULANCE CASE: WATER RESCUE 40.711839 -74.011234
5176 23:50:02 AMBULANCE CASE: WATER RESCUE 40.711839 -74.011234
5205 23:57:11 AMBULANCE CASE: UNCONSCIOUS/TRANSIT 40.732019 -74.000734
5211 23:57:59 AMBULANCE CASE: EDP/INSIDE 40.827547 -73.937461
5212 23:57:59 AMBULANCE CASE: EDP/INSIDE 40.827547 -73.937461
[459 rows x 4 columns]Let's add in the date and time features:
df = add_date_time_features(df)
print(df[['INCIDENT_DATE','WEEK_DAY','INCIDENT_TIME','INCIDENT_MIN']]) INCIDENT_DATE WEEK_DAY INCIDENT_TIME INCIDENT_MIN
7 07/04/2021 6 00:01:51 1.850000
27 07/04/2021 6 00:06:12 6.200000
51 07/04/2021 6 00:12:12 12.200000
53 07/04/2021 6 00:12:38 12.633333
54 07/04/2021 6 00:12:38 12.633333
... ... ... ... ...
5175 07/04/2021 6 23:50:02 1430.033333
5176 07/04/2021 6 23:50:02 1430.033333
5205 07/04/2021 6 23:57:11 1437.183333
5211 07/04/2021 6 23:57:59 1437.983333
5212 07/04/2021 6 23:57:59 1437.983333
[459 rows x 4 columns]
[Finished in 2.294s]WEEK_DAY column has the same value (i.e. 6) for every row.
Let's add in the date and time features:
df_early_am = filter_by_time(df,times=[0,360])
print(df_early_am[['INCIDENT_DATE','WEEK_DAY','INCIDENT_TIME','INCIDENT_MIN']]) INCIDENT_DATE WEEK_DAY INCIDENT_TIME INCIDENT_MIN
7 07/04/2021 6 00:01:51 1.850000
27 07/04/2021 6 00:06:12 6.200000
51 07/04/2021 6 00:12:12 12.200000
53 07/04/2021 6 00:12:38 12.633333
54 07/04/2021 6 00:12:38 12.633333
... ... ... ... ...
1041 07/04/2021 6 05:08:49 308.816667
1068 07/04/2021 6 05:21:49 321.816667
1075 07/04/2021 6 05:24:21 324.350000
1079 07/04/2021 6 05:28:13 328.216667
1111 07/04/2021 6 05:41:34 341.566667
[76 rows x 4 columns]
We can build a map with the calls for ambulances shaded by the time of the call:
import folium
import matplotlib.colors
def cc(minute,scale):
return(matplotlib.colors.to_hex( (minute/scale,0,minute/scale) ))
m = folium.Map(location=[40.7678,-73.9645],zoom_start=13,tiles="cartodbpositron")
df.apply( lambda row: folium.CircleMarker(location=[row["Latitude"], row["Longitude"]],
radius=5, popup=(row['INCIDENT_TIME']+": "+row['TYP_DESC']),
color=cc(row['INCIDENT_MIN'],2000))
.add_to(m) ,
axis=1)
m.save('4_July_map.html')We can also make maps with the computed clusters. We use the compute_locations function with different values of num_clusters. Since we are repeating the same actions for K = 2, 4, 6, we wrote a helper function to create the HTML maps:
def make_map(df, num_clusters, out_file):
centers,labels = compute_kmeans(df,num_clusters = num_clusters)
df_map = df[ ['Latitude','Longitude','INCIDENT_TIME','INCIDENT_MIN','TYP_DESC'] ]
df_map = df_map.assign(Labels = labels)
m = folium.Map(location=[40.7678,-73.9645],zoom_start=13,tiles="cartodbpositron")
df_map.apply( lambda row: folium.CircleMarker(location=[row["Latitude"], row["Longitude"]],
radius=5, popup=(row['INCIDENT_TIME']+": "+row['TYP_DESC']),
color=cc(row['Labels'],num_clusters))
.add_to(m), axis=1)
for i in range(num_clusters):
x,y = centers[i]
folium.Marker(location=[x,y],popup = "Cluster "+str(i)).add_to(m)
m.save(out_file)
make_map(df,2,'map_4_July_2clusters.html')
make_map(df,4,'map_4_July_4clusters.html')
make_map(df,8,'map_4_July_8clusters.html')Another function can be used to filter the dataset by day of the week and time of day. For example, still working with the 4 July data set:
df_mondays = filter_by_time(df, days = [0])
print(df_mondays)Empty DataFrame
Columns: [CAD_EVNT_ID, CREATE_DATE, INCIDENT_DATE, INCIDENT_TIME, NYPD_PCT_CD, BORO_NM, PATRL_BORO_NM, GEO_CD_X, GEO_CD_Y, RADIO_CODE, TYP_DESC, CIP_JOBS, ADD_TS, DISP_TS, ARRIVD_TS, CLOSNG_TS, Latitude, Longitude, WEEK_DAY, INCIDENT_MIN]
Index: []Let's next filter for early morning times:
df_early_am = filter_by_time(df,times=[0,360])
print(df_early_am[['INCIDENT_DATE','WEEK_DAY','INCIDENT_TIME','INCIDENT_MIN']]) INCIDENT_DATE WEEK_DAY INCIDENT_TIME INCIDENT_MIN
7 07/04/2021 6 00:01:51 1.850000
27 07/04/2021 6 00:06:12 6.200000
51 07/04/2021 6 00:12:12 12.200000
53 07/04/2021 6 00:12:38 12.633333
54 07/04/2021 6 00:12:38 12.633333
... ... ... ... ...
1041 07/04/2021 6 05:08:49 308.816667
1068 07/04/2021 6 05:21:49 321.816667
1075 07/04/2021 6 05:24:21 324.350000
1079 07/04/2021 6 05:28:13 328.216667
1111 07/04/2021 6 05:41:34 341.566667
[76 rows x 4 columns]We can use other methods for clustering, printing out the labels for the July 4 dataset:
print(compute_gmm(df))
print(compute_agglom(df))
[1 7 1 0 0 1 1 0 0 4 0 2 1 5 5 2 2 1 1 4 4 4 4 5 1 6 3 5 3 3 7 0 7 1 3 3 3
3 1 2 2 7 6 1 1 1 2 0 0 4 5 3 4 6 6 3 1 3 3 1 6 4 6 0 6 6 3 2 6 3 1 1 1 1
4 4 5 2 1 2 2 7 7 1 1 5 3 1 0 5 5 4 4 2 0 0 0 0 2 2 4 4 3 1 1 7 4 3 3 6 6
2 1 0 3 3 1 5 5 2 3 3 1 1 5 5 2 2 0 2 2 3 2 2 1 5 5 1 5 0 0 0 2 2 1 1 5 2
1 3 0 5 5 5 1 1 2 0 0 7 7 7 6 6 3 3 4 4 1 1 1 1 1 3 1 7 4 4 0 1 5 1 3 7 0
4 4 3 1 3 3 0 0 0 0 1 1 1 6 6 0 0 0 0 5 2 2 5 5 3 4 4 1 4 2 0 2 0 2 2 1 1
1 3 3 1 3 3 1 0 0 5 5 1 5 5 1 7 5 2 1 0 0 0 1 5 2 0 2 1 4 4 1 1 4 7 7 3 3
4 1 1 7 7 5 5 0 1 5 1 1 0 7 0 0 2 0 7 7 5 1 2 3 1 1 1 1 3 6 6 4 5 5 2 2 0
3 0 0 2 5 5 0 1 3 7 7 5 5 3 0 6 0 0 1 2 4 5 2 1 3 3 1 1 2 0 1 2 2 2 3 3 4
4 5 5 1 2 0 3 4 3 3 0 1 6 1 1 4 1 4 4 3 6 6 6 2 2 2 3 6 3 3 1 0 5 3 5 2 2
3 3 6 6 2 2 3 6 6 2 0 0 3 3 2 0 3 2 2 3 3 4 4 6 6 1 6 4 5 5 5 4 6 1 1 1 4
2 0 3 5 2 1 3 1 4 3 1 0 3 5 5 5 3 3 7 3 1 2 2 2 4 4 2 2 2 1 1 3 3 2 3 5 1
1 1 0 0 0 0 4 4 5 6 6 6 1 2 2]
[0 6 0 3 3 0 0 3 3 2 3 4 0 7 7 4 4 0 0 2 2 2 2 7 0 0 1 0 1 1 6 3 1 0 1 1 1
1 0 4 4 4 5 0 0 0 4 3 3 2 3 1 2 0 0 1 0 1 1 0 0 2 0 3 0 0 1 4 0 1 0 0 0 0
2 2 3 4 0 4 4 1 1 1 0 3 1 0 3 7 7 2 2 4 3 3 3 3 4 4 2 2 1 0 0 6 2 1 1 0 0
4 0 4 1 1 0 0 0 4 1 1 0 0 7 7 4 4 3 4 4 1 4 4 0 7 7 0 7 3 3 3 4 4 0 0 7 4
0 1 3 7 0 0 0 0 4 3 3 6 6 4 5 5 1 1 2 2 0 0 0 0 0 1 0 6 2 2 3 0 7 0 1 6 3
2 2 1 0 0 0 3 3 3 3 0 0 0 0 5 3 3 3 3 7 4 4 7 7 1 2 2 0 4 4 3 4 6 4 4 0 0
0 1 1 0 1 1 0 3 3 7 7 0 3 3 0 6 3 4 0 3 3 3 0 7 4 3 4 0 2 4 0 0 2 6 6 1 1
2 0 0 6 6 3 3 3 0 7 5 5 3 6 3 3 4 3 6 6 0 0 4 1 0 0 0 0 1 5 5 2 7 7 4 4 3
1 3 3 4 7 7 3 0 1 6 6 7 7 7 3 0 3 3 0 4 2 7 4 0 1 1 1 1 4 3 0 4 4 4 1 1 2
2 0 0 0 4 3 1 2 1 1 3 0 5 0 0 2 0 2 2 1 5 5 5 4 4 4 1 5 1 1 0 3 7 1 3 4 4
1 1 5 5 4 4 1 0 0 4 3 3 1 1 4 3 1 4 4 1 1 2 2 5 5 0 5 2 7 3 3 2 5 0 0 0 2
4 4 1 7 4 0 1 0 2 1 0 3 1 7 3 3 1 1 6 1 0 4 4 4 2 2 4 4 4 0 0 1 1 4 1 3 0
0 0 3 3 3 3 2 2 3 5 5 5 0 4 4]
Lastly, we can use the function compute_explained_variance to assess the best number of clusters:
k_vals = list(range(1,20))
ev = compute_explained_variance(df,K=k_vals)
sns.lineplot(k_vals,ev)
plt.title('Explained Variance for KMeans for Manhattan, 4 July 2021')
plt.show()

showing a sharp drop-off to K=5 that quickly flattens, showing that additional clusters, beyond 10, do not significantly improve the average distance to the assigned means. This suggests that a reasonable number of clusters is around 8.