
You can check if your answers are right, but typing them into a Python shell.
blurb = "Welcome to MHC, the Honors College at CUNY. MHC provides the transformational experiences that will take students from their academic aspirations into careers as leaders in their chosen fields. We have already seen that transformation taking place, as more of our accomplished young alumni make their mark on NYC and the world. Nothing is more satisfying to me than being able to positively impact the lives of NYC's most promising undergraduate students. I invite you to learn more about MHC and our remarkable students and alumni at MHC, CUNY. Attend an open house or cultural event, read some recent press or browse this site.
Mary C. Pearl, Ph.D."
"POINT (40.715 -73.99)"and extracts out the two numbers 40.715 and -73.99.
Regular expressions provide a powerful way to find patterns, in particular, those that might vary in length. Patterns can be as simple as a single word, or a series of strings that can occur a fixed or varying number of times. For example, if you were searching for any number of the letter a, you could write:
a*which says you are looking for 0 or more a. Similarly, if you wanted a word repeated:
(hi)*This pattern will match any number of copies of the word hi, such as: hi, hihihihihi, etc.
This search for patterns are quite useful in many fields including biology (yup, this was added just for all the biology majors in the class). For example, in a DNA sequence, small patterns can occur varying number of times (short tandem repeat polymorphism). To find the first AT repeat of longer than 4 repeats, we can use a regular expression:
import re
dna = "ACTGCATTATATCGTACGAAAGCTGCTTATACGCGCG"
runs = re.findall("[AT]{4,100}", dna)
print(runs)
To find the location of a pattern in a string, we can use:
if re.search(r"GC[ATGC]GC", dna):
print("restriction site found!")
(for the biologists in the class: more examples like this from Python for Biologists).
The re library is distributed with Python. We will use two useful functions in the library:
We often want more information than just if a pattern occurred or in what way. To find out the starting (and stopping) location, we can use the match object that re.search() returns. It's most useful functions are:
From our example above, we could store the match object:
m = re.search(r"GC[ATGC]GC, dna) print "The matching string is", m.group() print "Match starts at", m.start() print "Match ends at", m.end()
These are more general (and more powerful) tools than the string methods above. In many cases, either can be used. For finding approximate matches or matches of varying lengths, using regular expressions is much easier. Here's a regex cheat sheet with an overview of the most common commands.
dna = "ACTGCATTATATCGTACGAAAGCTGCTTATACGCGCG"
College or Institution Type Campus Campus Website Address City State Zip Latitude Longitude Location Senior Colleges Baruch College http://baruch.cuny.edu 151 East 25th Street New York NY 10010-2313 40.740977 -73.984252 (40.740977, -73.984252) Senior Colleges Brooklyn College http://brooklyn.edu 2900 Bedford Avenue Brooklyn NY 11210-2850 40.630276 -73.955545 (40.630276, -73.955545) Community Colleges Borough of Manhattan Community College http://bmcc.cuny.edu 199 Chambers Street New York NY 10007-1044 40.717367 -74.012178 (40.717367, -74.012178) Community Colleges Bronx Community College http://bcc.cuny.edu 2155 University Avenue Bronx NY 10453 40.856673 -73.910127 (40.856673, -73.910127) Senior Colleges The City College of New York http://ccny.cuny.edu 160 Convent Avenue New York NY 10031-9101 40.819548 -73.949518 (40.819548, -73.949518) Graduate Colleges CUNY School of Law http://law.cuny.edu 2 Court Square Long Island City NY 11101-4356 40.747639 -73.943676 (40.747639, -73.943676) Graduate Colleges The Graduate School and University Center http://gc.cuny.edu 365 5th Avenue New York NY 10016-4309 40.748724 -73.984205 (40.748724, -73.984205) Senior Colleges Hunter College http://hunter.cuny.edu 695 Park Avenue New York NY 10065-5024 40.768731 -73.964915 (40.768731, -73.964915)Hint: Think about the pattern a zip code has.
Webpages are formatted using the HyperText Markup Language (HTML) which is a series of commands on how the pages should be formatted, along with links and embedded calls to programs. For example, if you would like a word to show up in bold, you surround it by "tags" that say what to do:
<b>bold</b>The opening tag starts the bold text and the closing tag (indicated by the '/') ends the bold text. Most HTML commands follow this same style: there's an opening tag, paired with a closing text that includes a '/' and the same name.
We can access files stored on webpages inside Python. The built-in urllib module has functions to mimic the file I/O we have seen before. If we are reading in a CSV file, we can use pandas directly (see the citiBike example in Classwork 9).
Let's say we want to make a list of all the seminars at the American Museum of Natural History (we're using these, since I like to go to their seminars, and the format is a bit easier than the MHC events page which is scattered across multiple pages). We can `scrap the data' on the comparative biology seminar page into our Python program. We can then search for strings that look like dates and print out the subsequent lines. The interface is very similarly to opening files:
The museum's webpage is machine generated (you can look at the raw code by saving the HTML file and then opening it with TextEdit, TextWrangler, or some other editor meant for text files). The code is very clean with no missing ending tags (unlike the HTML for the page you're currently reading...).
Here are the first couple of lines with the seminar dates:
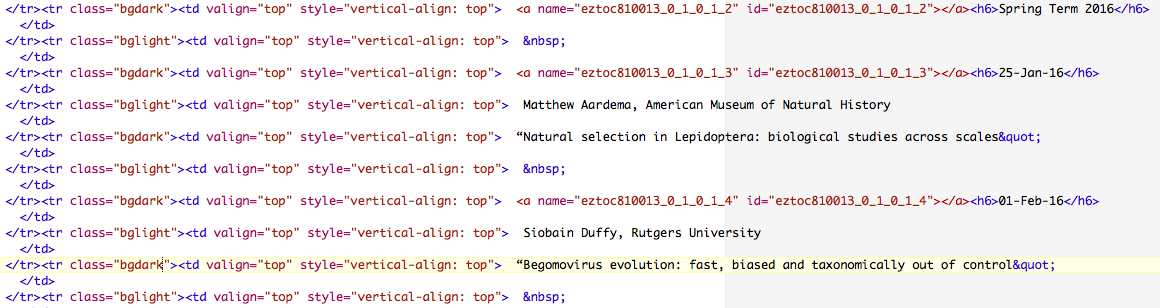
We can search the file for dates, and then print out the subsequent lines with the speaker and title. We can do this in several different ways. Here's one approach:
We are just missing the tools to open webpages. There are several options (both built-in and modules you can download). We are going to use requests since it automatically converts incoming data from bytes to text, making it simplier to use. First we need to import the module:
import requests
If you do not have requests on your machine, you can download it in a terminal with:
pip install requests
To get the contents of a webpage:
data = requests.get("http://www.amnh.org/our-research/richard-gilder-graduate-school/academics-and-research/seminars-and-conferences")
which now contains the strings in the file as well as some other information (This will take a bit depending on network connectivity.) The text of the file can be accessed via:
data.text
It's stored as a single, very long string with \n separating the lines. So, if we want to look at the file line-by-line, we can use our friend, split()
lines = data.text.split("\n")
If you print out lines, you'll notice that most lines are blank or formatting statements. To find the seminars, we need to go through and check each line to see if it contains a seminar listing (hint: if statement!).
Since each line of the webpage is in the variable lines, and we can loop through it. Here's an outline: it traverses the list by line number since we'll want to refer to the lines after it (where the name and titles are stored):
for i in range(len(lines)): #Check if the lines[i] has a date in it (can use find() or regular expressions) #If it does print it, # as well as the subsequent lines[i+2] (has name) and # lines[i+4] (has title)
Test and debug your program and then figure out how to print just the date, name, affiliation, and title (without the HTML formatting statements).
For those who are going to be digging deep into webpages, there's a lovely package that makes it much easier (if the above web scraping was more than enough for you, skip this section).
The Beautiful Soup package for Python has the motto:
You didn't write that awful page. You're just trying to get some data out of it. Beautiful Soup is here to help. Since 2004, it's been saving programmers hours or days of work on quick-turnaround screen scraping projects.
Like folium or pandas, you will need to download the package to your machine to use it. Here is the soup quick start.