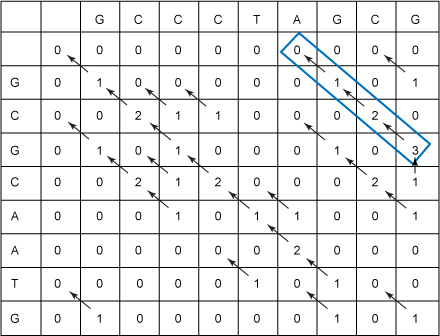
Dynamic Programming
For problems that can be broken into subproblems (and those subproblems occur mulitple times), dynamic programming is often a good approach to compute the answer quickly. The textbook introduces the Manhattan Tourist Problem, which we will implement, and then use the framework developed for it to implement the Needleman-Wunsch algorithm for global pairwise sequence alignment.Implementing the Manhattan Tourist:
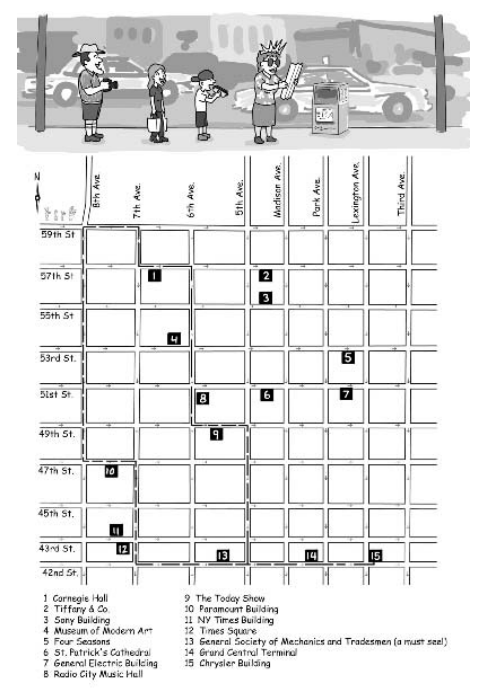
The textbook introduces the Manhattan Tourist problem, where you have a grid of streets with landmarks, and you would like to see as many as possible, without retracing your steps. For example, you start at Columbus Circle (59th Street and 8th Avenue) and finish at 42nd Street and 3rd Avenue:
We can apply this approach to an arbitrarily labeled grid (assume the upper corner has coordinates (0,0) and each increases and the coordinates increase as you progress south and east:
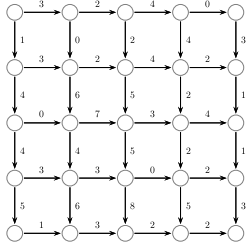
The steps we outlined were:
- Store best path so far in a grid (array).
- Fill in the first row and column separately (since there's only one direction from which to reach those cells)
- Fill in the second row (using the first).
- Fill in the third row (using the second).
- Repeat until all rows are filled.
- The most number of landmarks you can visit is in the bottom right cell.
To do this, we will need:
- south: number of landmarks seen going south from a location.
- east: number of landmarks seen going east from a location.
- best: an array to store the largest possible number of landmarks seen by the time you reach a location.
import numpy as np #The number of landmarks going east and south from a location: east = np.array( [[3,2,4,0],[3,2,4,2],[0,7,3,4],[3,3,0,2],[1,3,2,2]] ) south = np.array( [[1,0,2,4,3],[4,6,5,2,1],[4,4,5,2,1],[5,6,8,5,3]] ) size = 5 best = np.zeros( (size,size) ) #Check to make sure this matches figure: print "east:", east print "south:", south
Now, let's fill out first row and column of the grid:
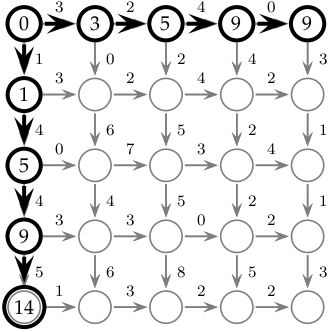
so that the upper corner is 0, and the elements in the first column depend just on the elements above them, and similarly, the elements in the first row depend on those to their left:
best[0,0] = 0
for i in range(1,size):
best[0,i] = best[0,i-1] + east[0,i-1]
best[i,0] = best[i-1,0] + south[i-1,0]
#Check that it works:
print best
To fill in the rest of the array, we use the larger of the total seen entering going east and entering going south:
#Fill in the rest of the array:
for i in range(1,size):
for j in range(1,size):
best[i,j] = max(best[i,j-1] + east[i,j-1], best[i-1,j] + south[i-1,j])
#Check that it works:
print best
Once we check that the above works, then we can add in the code that "reads off" the path from computatations. The easiest way to do this is to create a second matrix, traceback that keeps track of the direction from which the best path enters each element. To the top of the file, let's add code that creates a 2D array of dimensions (size,size) to hold the traceback. numpy arrays only take numbers, but when figuring out the path, it is easier to think of words "UP" and "LEFT" (then numbers. To make the code easier to read, we will create variables called UP = 1 and LEFT = -1 and use those:
traceback = np.zeros( (size,size) ) UP = 1 LEFT = -1
Now, when we compute the values for best, let's also keep track of where they came:
#Set up first row and first column to gap:
best[0,0] = 0
for i in range(1,size):
best[0,i] = best[0,i-1] + east[0,i-1]
best[i,0] = best[i-1,0] + south[i-1,0]
traceback[i,0] = UP
traceback[0,i] = LEFT
print traceback
#Fill in the rest of the array:
for i in range(1,size):
for j in range(1,size):
best[i,j] = max(best[i,j-1] + east[i,j-1], best[i-1,j] + south[i-1,j])
if best[i,j-1] + east[i,j-1] > best[i-1,j] + south[i-1,j]:
traceback[i,j] = LEFT
else:
traceback[i,j] = UP
print traceback
Once we have the traceback, we can use it to create the path. We'll start at the lower left corner (i.e. (i,j) = (size-1,size-1)) and then we write down the value we stored in the traceback for it, go to that point, write down its value, and repeat until we reach the origin:
path = ""
i = size-1
j = size-1
while i > 0 or j > 0:
if traceback[i,j] == UP:
path = "south " + path
i = i - 1
else:
path = "east " + path
j = j -1
print path
Check to make sure that your path is correct before continuing to sequence alignment.