Program 11: Digit Dimensions
CSci 39542: Introduction to Data Science
Department of Computer Science
Hunter College, City University of New York
Spring 2022
Classwork Quizzes Homework Project
Program Description
Program 11: Digit Dimensions. Due noon, Thursday, 29 April.
In Lecture 22 and Chapter 26, we applied Principal Components Analysis as a dimensionality reduction technique and used scree plots to provide a visualization of the captured variance. This assignment continues our analysis of the sklearn digits dataset, focusing on visualization and intrinsic dimenion of the data.
In addition, we will write a function that allows the user to explore how many dimensions are needed to see the underlying structure of images from the sklearn digits dataset (inspired by Python Data Science Handbook: Section 5.9 (PCA)).
As in Program 10, we can view our images as "flattened" 1D arrays of length 64.
As discussed in Python Data Science Handbook: Section 5.9, we can view the images as sums of the components.
If we let
In a similar fashion, we can represent the image in terms of the axis,
In your program, include the following functions from Program 10. You may use your earlier function or the Program 10 solution available on Blackboard:
Learning Objective: to increase facility with standard linear algebra approaches and strengthen understanding of intrinistic dimensions of data sets via exploration of the classic digits dataset).
Available Libraries: pandas, numpy, pickle, sklearn, and core Python 3.6+.
Data Sources: MNIST dataset of hand-written digits, available in sklearn digits dataset.
Sample Datasets: sklearn digits dataset.
x1 = [1 0 ... 0],
x2 = [0 1 0 ... 0], ...,
x64 = [0 ... 0 1] (vectors corresponding to the axis), then we can write our images, im = [i1 i2 ... i64], as: im = x1*i1 + x2*i2 + x3*i3 + ... + x64*i64.
For example, if we take the first entry from the dataset:
We can write it as:
[ 0. 0. 5. 13. 9. 1. 0. 0. 0. 0. 13. 15. 10. 15. 5. 0. 0. 3. 15. 2. 0. 11. 8. 0. 0. 4. 12. 0. 0. 8. 8. 0. 0. 5. 8. 0. 0. 9. 8. 0. 0. 4. 11. 0. 1. 12. 7. 0. 0. 2. 14. 5. 10. 12. 0. 0. 0. 0. 6. 13. 10. 0. 0. 0.]
plugging in the values for a given image (in this case the first image in the dataset) into the equation.
im = x1*i1 + x2*i2 + x3*i3 + ... + x64*i64
= x1*0 + x2*0 + x3*5 + ... + x64*0c1, c2, ... c64, that the PCA analysis returns:
since the axis of PCA are chosen so that the first one captures the most variance, the second the next most, etc. The later axis capture very little variance and likely add litte to the image. For technical reasons, we include the mean (the reason is similar to when we "center" multidimensional data at 0).
This can be very useful for reducing the dimension of the data set, for example, here is the first image from above on the left:
im = mean + c1*i1 + c2*i2 + ... + c64*i64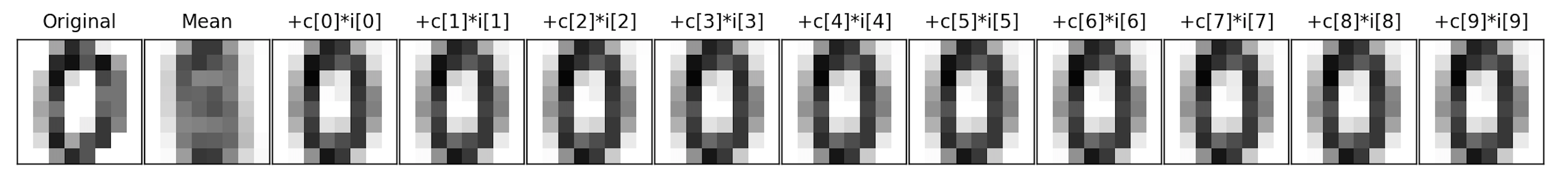
The next image is the overall mean, and each subsequent image is adding another component to the previous. For this particular scan, the mean plus its first component is enough to see that it's a 0.
And write the following new functions:
select_data(data, target, labels = [0,1]):
This function takes as three input parameters:
Returns the rows of data: a numpy array that
includes rows of equal size flattened arrays,
target a numpy array that contains the labels for each row in data.
labels: the labels from target that the rows to be selected. The default value is [0,1].
data and
target where the value of target is in labels.
run_pca(xes):
This function takes as one input parameter:
It fits a model of sklearn.decomposition.PCA to the
xes: a numpy array that
includes rows of equal size flattened arrays,
xes. The function returns the model and the transformed values.
capture_85(mod):
This function takes as one input parameter:
From the array of the amount of varianace of each component, (stored in mod: a model of sklearn.decomposition.PCA that has been fitted to a dataset.
mod.singular_values_),
compute fraction of the variance of each component. That is, if sv = mod.singular_values_, then s=sv**2/sum(sv**2) (see Chapter 26.3.1.1 Captured Variance and Scree Plots).
Returns the number of elements needed to capture more than 85% of the variance.
For example, if sv is:
then array([585.57, 261.06, 166.31, 57.14, 48.16, 39.79, 31.71, 28.91,
24.23, 22.23, 20.51, 18.96, 17.01, 15.73, 7.72, 4.3 ,
1.95, 0.04])s is
The first component captures 76% of the variance which is lower than that 85% threshold. The first and second components capture (76+15)% = 91% of the variance, so, your function would return 2.
array([0.76, 0.15, 0.06, 0.01, 0.01, 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. ])average_eigenvalue(mod):
This function takes as one input parameter:
For the mod: a model of sklearn.decomposition.PCA that has been fitted to a dataset.
mod,
computes the average of the eigenvalues (i.e. the average of sv = mod.singular_values_) and returns the number of elements greater than the average. See sklearn.decomposition.PCA for attributes of the model.
approx_digits(mod, img, numComponents=8):
This function has three inputs:
The function transforms the image, mod: a model of sklearn.decomposition.PCA that has been fitted to a dataset.
img: a flattened image from the dataset.
numComponents: the number of components used in the approximation. Expecting a value between 0 and 64. The default value is 8.
img, and uses the resulting coefficents to compute an approximation of the image. The approximation image (flattened array) is the mean plus the sum of the first numComponents
terms (i.e. coefficients[i] * components[i] where the coefficients are returned from the transform and components are an array of the components computed by PCA() analysis).
Note that the mean of the dataset does not have to be computed since it is stored as an attribute of the models of PCA().
Returns the computed approximation image as a flattened array.
Hint: see the discussion in DS Handbook 5.09 (PCA) and in Lecture 21.
Let's run through a few examples. We can start with our binary digits from Program 10:
from sklearn import datasets
digits = datasets.load_digits()
n_samples = len(digits.images)
data = digits.images.reshape((n_samples, -1))
target = digits.target
print(f'The targets for the first 5 entries: {target[:5]}')
bin_dig, bin_tar = select_data(data,digits.target)
print(f'The targets for the first 5 binary entries: {bin_tar[:5]}')pca = PCA().fit(digits.data)
The targets for the first 5 binary entries: (0, 1, 0, 1, 0)Using our function from Program 10, we can select the binary digits and then fit the PCA model to those. Our function returns the model as well as the transformed values:
import matplotlib.pyplot as plt
bin_data,bin_target = select_data(data,target)
bin_mod, bin_proj = run_pca(bin_data)
scatter = plt.scatter(bin_proj[:, 0], bin_proj[:, 1], c=bin_target, alpha=0.5,
cmap=plt.cm.get_cmap('rainbow', 10))
plt.title('PCA for digits')
plt.xlabel('Component 1')
plt.ylabel('Component 2')
plt.legend(*scatter.legend_elements())
plt.show()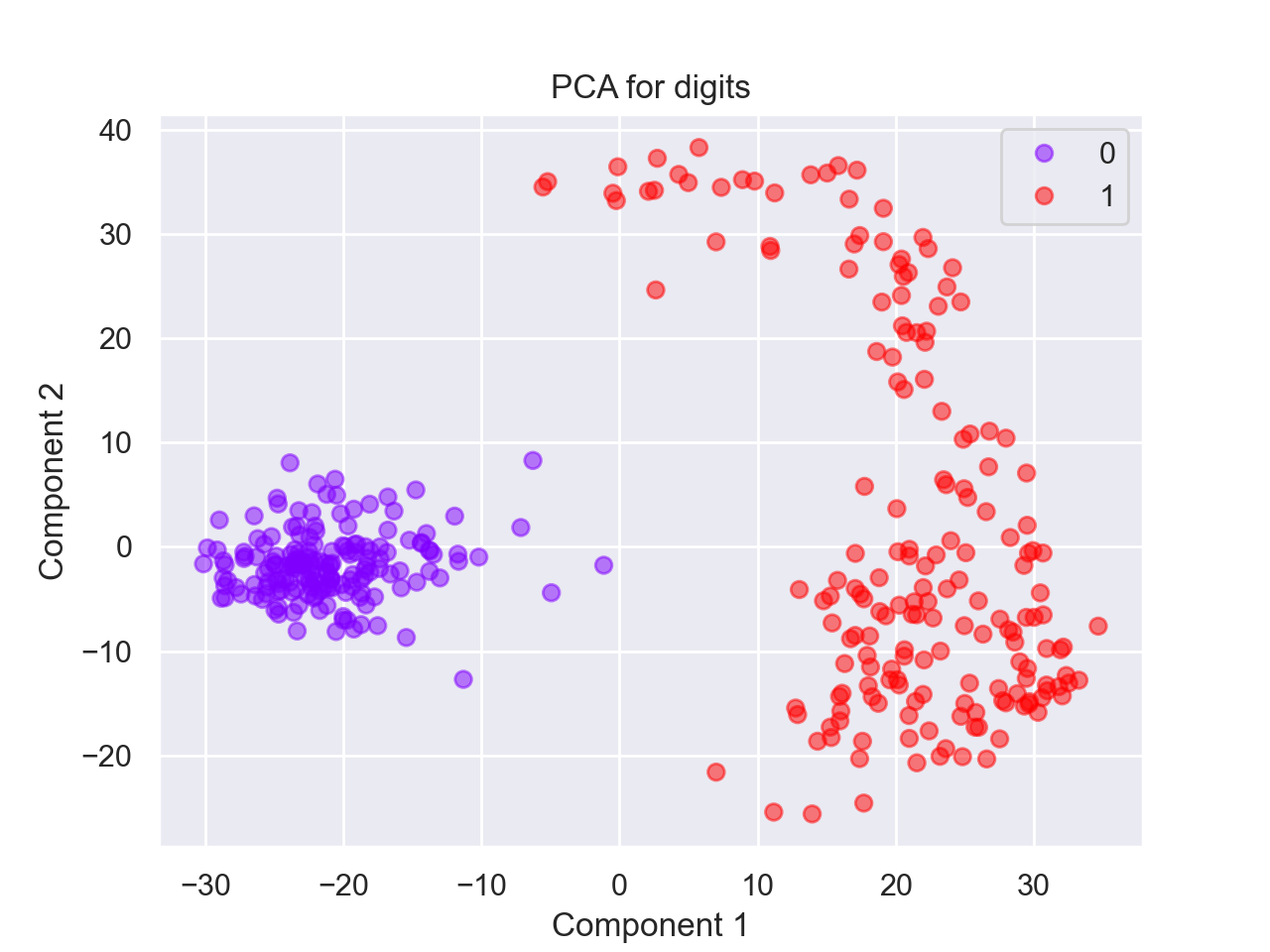
We can fit the model to all the digits:
all_mod, all_proj = run_pca(data)
scatter = plt.scatter(all_proj[:, 0], all_proj[:, 1], c=digits.target, alpha=0.5,
cmap=plt.cm.get_cmap('rainbow', 10))
plt.title('PCA for digits')
plt.xlabel('Component 1')
plt.ylabel('Component 2')
plt.legend(*scatter.legend_elements())
plt.show()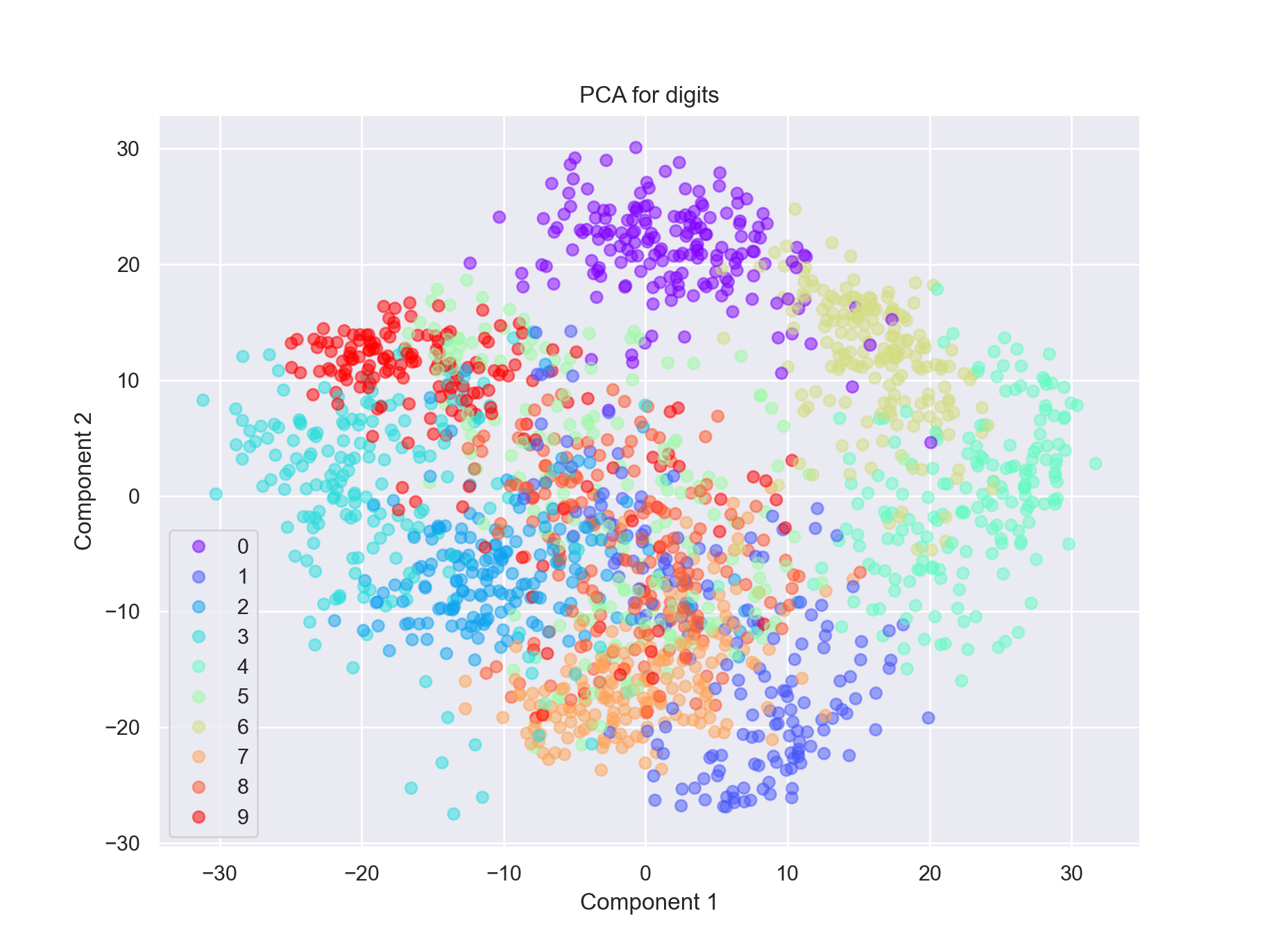
Plotting the explained variance ratio of our model:
import numpy as np
plt.plot(np.cumsum(all_mod.explained_variance_ratio_))
plt.xlabel('number of components')
plt.ylabel('cumulative explained variance')
plt.show()
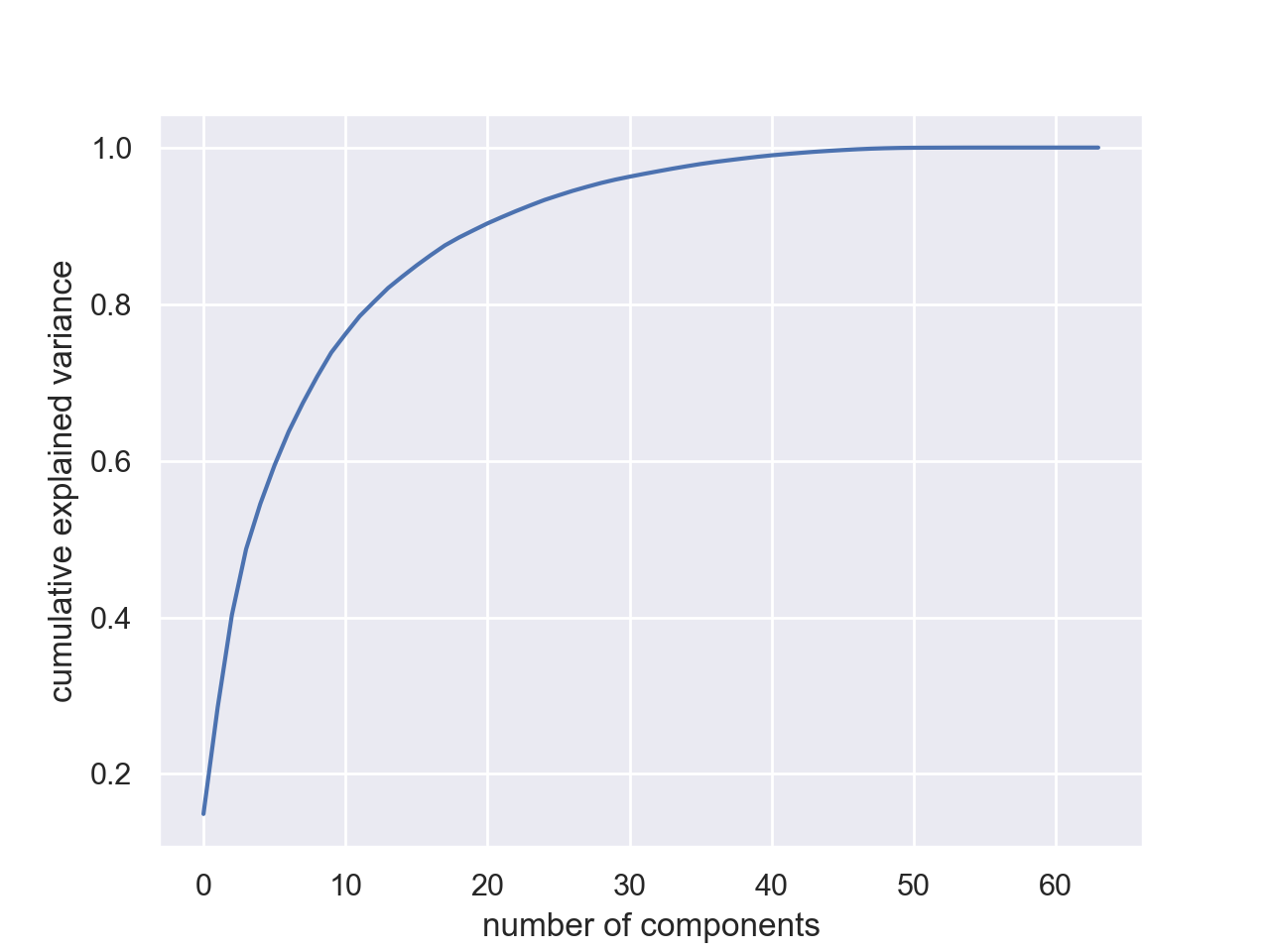
We can use our functions to look at ways to measure this, via the captured variance and average eigenvalues:
np.set_printoptions(suppress=True) #Turn off scientific notation
sv = all_mod.singular_values_
print(f'The singular values are: {sv}')
print(f'The fraction variance: {sv**2/sum(sv**2)}')
print(f'The number of components needed to capture 85% of the variance is {answer.captures85(all_mod)}.')
print(f'The number of eigenvalues larger than the average is {answer.averageEigenvalue(all_mod)}.')
The singular values are: [567.0065665 542.25185421 504.63059421 426.11767608 353.3350328
325.82036569 305.26158002 281.16033073 269.06978193 257.82395143
226.31879719 221.5148324 198.33071545 195.70013887 177.9762712
174.46079067 168.72787641 164.15849219 148.23330876 139.83132462
138.58443271 131.1882069 128.72691665 124.93159016 122.57503405
113.44487728 111.48027133 105.46348813 102.80780243 96.22856616
89.81296469 87.33494649 85.25960437 84.15671337 81.58936529
79.64200462 74.43047136 70.12195688 69.27559227 67.56406817
64.03315896 58.52697795 57.12818557 55.09243185 50.17909986
48.1749428 45.62286487 40.89585718 34.68503519 29.5461187
21.28899661 13.34460261 10.64814019 10.44437712 8.44041164
5.18181553 3.90097916 2.55109635 1.51445826 1.08979014
0.86043771 0. 0. 0. ]
The fraction variance: [0.14890594 0.13618771 0.11794594 0.08409979 0.05782415 0.0491691
0.04315987 0.03661373 0.03353248 0.03078806 0.02372341 0.02272697
0.01821863 0.01773855 0.01467101 0.01409716 0.01318589 0.01248138
0.01017718 0.00905617 0.00889538 0.00797123 0.00767493 0.00722904
0.00695889 0.00596081 0.00575615 0.00515158 0.0048954 0.00428888
0.00373606 0.00353274 0.00336684 0.0032803 0.00308321 0.00293779
0.00256589 0.00227742 0.00222278 0.0021143 0.00189909 0.00158653
0.0015116 0.00140579 0.00116622 0.00107493 0.00096405 0.00077463
0.00055721 0.00040433 0.00020992 0.00008248 0.00005251 0.00005052
0.000033 0.00001244 0.00000705 0.00000301 0.00000106 0.00000055
0.00000034 0. 0. 0. ]
The number of components needed to capture 85% of the variance is 17.
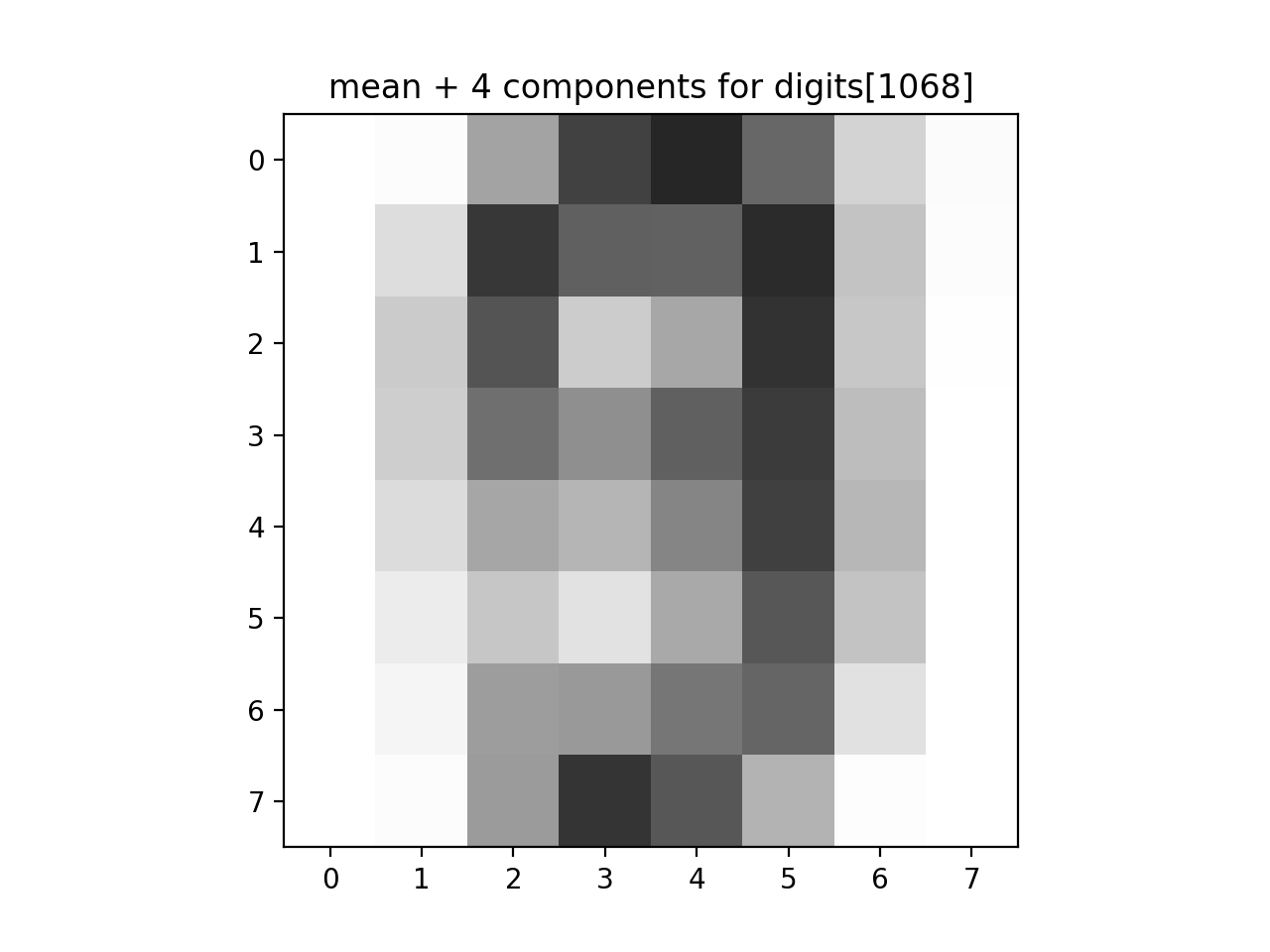
The number of eigenvalues larger than the average is 23.Lastly, let's look at how many components are needed to recover a given image:
plt.imshow(all_mod.mean_.reshape(8, 8), cmap='binary', interpolation='nearest', clim=(0, 16))
plt.title("Mean for digits")
plt.show()
print(f'The original image: {data[1068]}.')
approx_answer = approx_digits(all_mod, all_proj[1068], numComponents=4)
print(f'The first 4 components: {approx_answer}')
plt.imshow(approx_answer.reshape(8, 8), cmap='binary', interpolation='nearest', clim=(0, 16))
plt.title("mean + 4 components for digits[1068]")
plt.show()
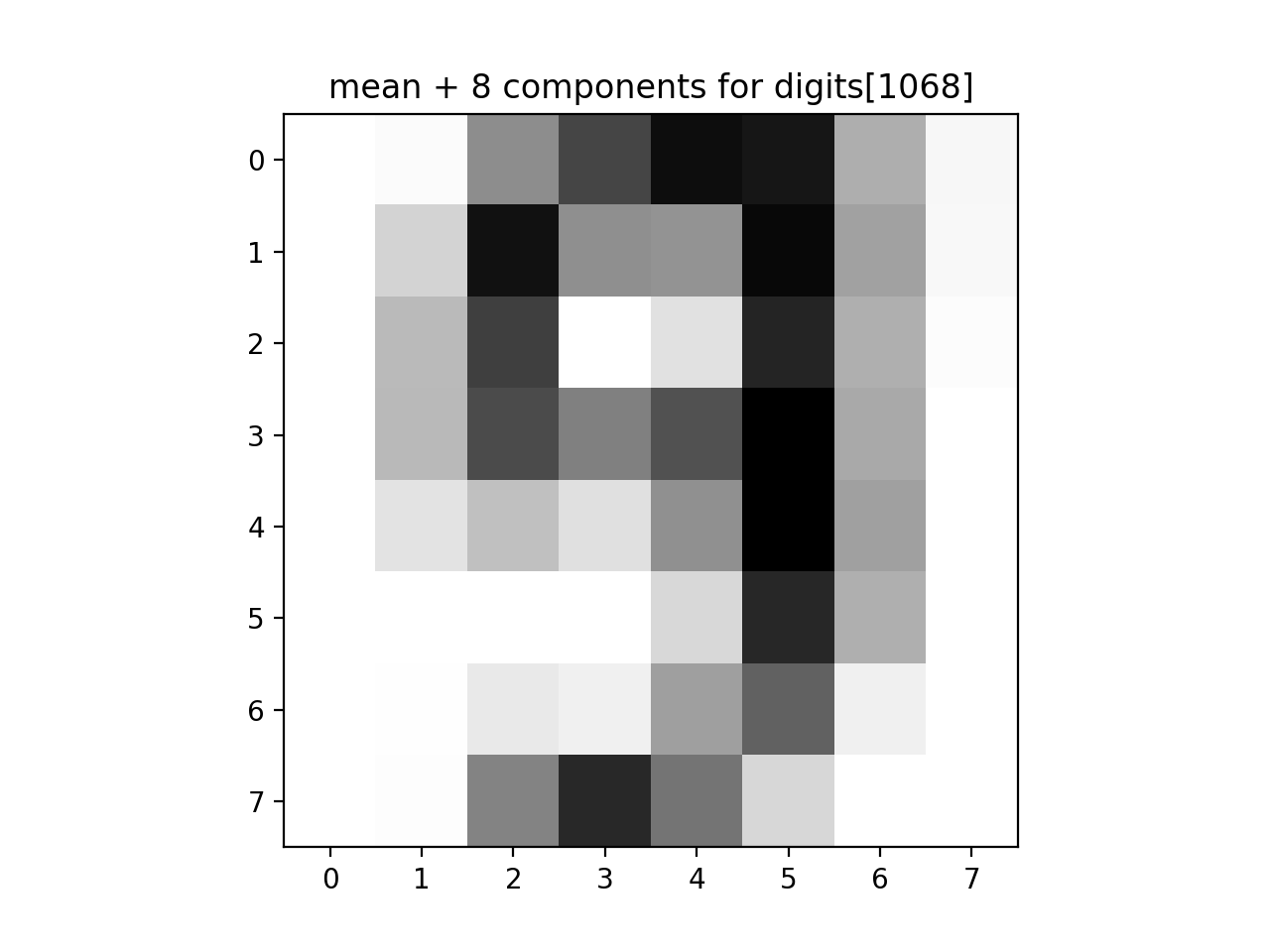
approx_answer = approx_digits(all_mod, all_proj[1068], numComponents=8)
print(f'The first 8 components: {approx_answer}')
plt.imshow(approx_answer.reshape(8, 8), cmap='binary', interpolation='nearest', clim=(0, 16))
plt.title("mean + 8 components for digits[1068]")
plt.show()
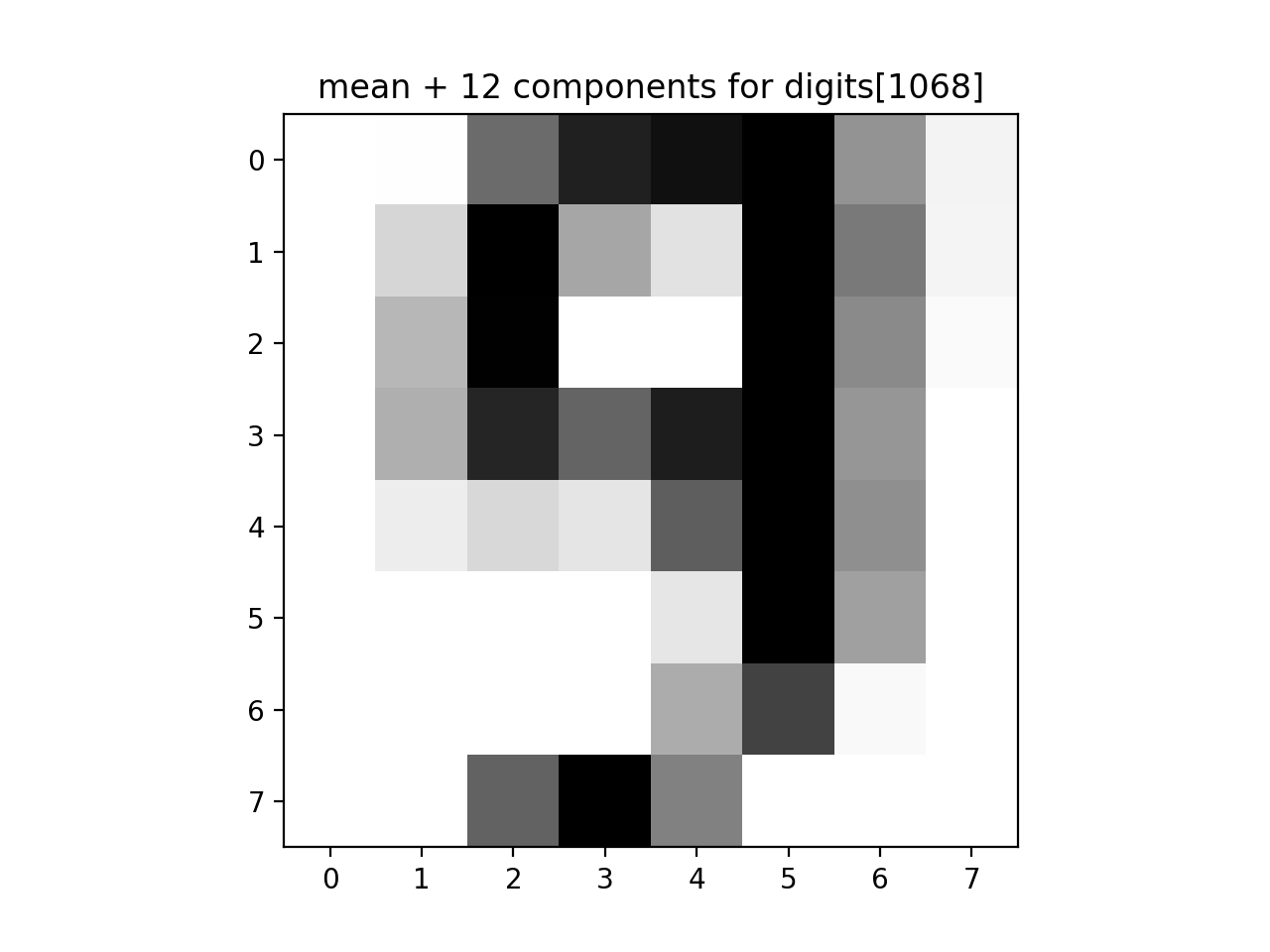
approx_answer = approx_digits(all_mod, all_proj[1068], numComponents=12)
print(f'The first 12 components: {approx_answer}')
plt.imshow(approx_answer.reshape(8, 8), cmap='binary', interpolation='nearest', clim=(0, 16))
plt.title("mean + 12 components for digits[1068]")
plt.show()data[1068]: